Procedural steps
This protocol paper 1 was a very good resource for understanding the procedural steps involved in any RNA-Seq analysis. The datasets they use in that paper are freely available, but the source of RNA was the fruitfly Drosophila melanogaster, and not Human tissue. In addition, they exclusively use the “tuxedo” suite developed in their group.
Several papers are now available that describe the steps in greater detail for preparing and analyzing RNA-Seq data, including using more recent statistical tools:
In addition, newer alignment-free methods have also been published and are increasingly being used in analysis (we include a second protocol detailing the use of these):
The sections below detail those protocols and suggest tools.
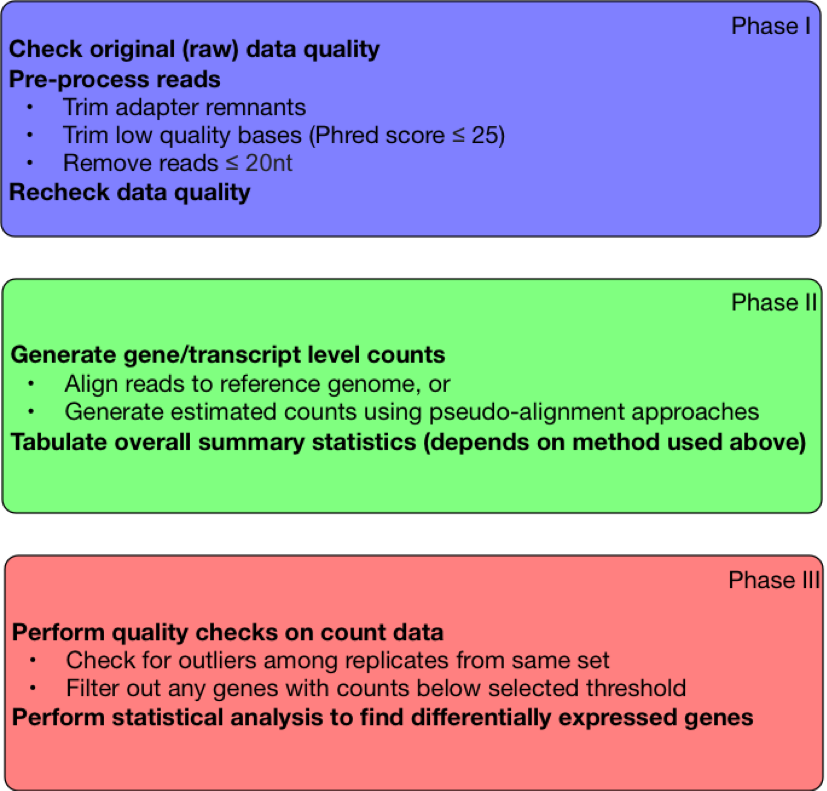 |
|---|
| Figure 1. Steps in RNA-Seq Workflow |
Bibliography
-
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., … & Pachter, L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature protocols, 7(3), 562. ↩
-
Love MI, Anders S, Kim V and Huber W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Research 2016, 4:1070. ↩
-
Law CW, Alhamdoosh M, Su S et al. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Research 2018, 5:1408. ↩
-
Conesa, A., Madrigal, P., Tarazona S., … & Mortazavi, A. 2016. A survey of best practices for RNA-Seq data analysis. Genome Biology 17:13. https://doi.org/10.1186/s13059-016-0881-8 ↩
-
Bray, N. L., Pimentel, H., Melsted, P., & Pachter, L. (2016). Near-optimal probabilistic RNA-seq quantification. Nature biotechnology, 34(5), 525. ↩
-
Patro, R., Duggal, G., Love, M. I., Irizarry, R. A., & Kingsford, C. (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nature methods, 14(4), 417. ↩

This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 4.0 International License.